ES cells and iPS cells are ideal models for analyzing physiological phenomena unique to stem cells, such as maintenance of pluripotency and cell differentiation. In our laboratory, we analyze these dynamic phenomena by making full use of molecular biology, cell biology, and genome science. In particular, we focus on the forward genetic approach; we are actively exploring unknown gene functions that cannot be predicted.
1. Analysis of Pluripotency Regulation Mechanism by Forward Genetics Through the Use of Homozygous Mutant Mouse ES Cell Lines
In forward genetics, a large number of mutants are generated, and then those with the desired phenotype are identified. Because no assumption is made about the function of the gene when the mutants are generated, it is highly likely that the newly revealed gene functions have not been anticipated by researchers. In fact, in model organisms such as E. coli and yeast, many breakthroughs have been achieved through forward genetics. In mammalian cells, however, completely disrupting gene functions is not easy because each gene has two alleles. To challenge this problem, we have developed a technique to rapidly produce homozygous mutant cells (Horie, Nature Methods 2011, Figure 1, Movie 1). In this method, heterozygous mutants are generated by using retroviruses or transposons as mutagens. Then, by using a tetracycline system to transiently suppress the Bloom gene, we induce recombination between homologous chromosomes, thus obtaining homozygous mutants from heterozygous mutants.
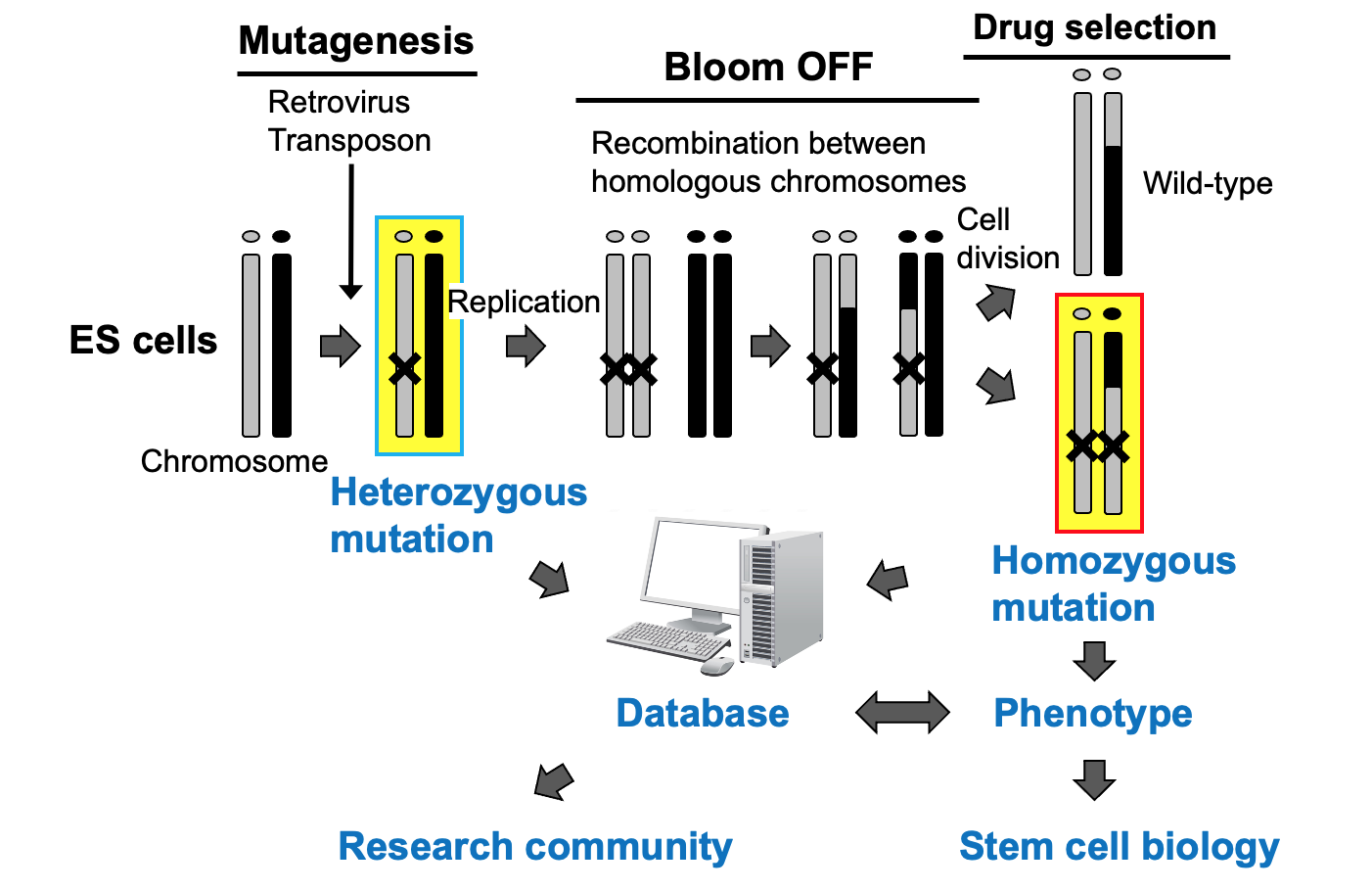
Figure 1. Generation of homozygous mutant ES cells.

Movie 1. Genatation of homozygous mutants by controlling the Bloom gene
We have generated about 2,000 heterozygous mutant ES cell lines and about 200 homozygous ES cell lines, and we have identified mutations that exhibit a variety of phenotypes. By comparing and analyzing a large number of mutants, we are trying to gain a comprehensive understanding of ES cell pluripotency and have discovered a new biological pathway that regulates pluripotency (Yoshida, Stem Cell Reports 2025; Kashiwagi, J Cell Sci 2024). These mutant ES cells have been registered in international databases and are being used by other researchers (Nakai, Nat Commun 2024; Umemura, PLoS One 2013). We have also reported the genome insertion mechanisms of retroviruses and transposons to improve the efficiency of genome mutagenesis (Yoshida, Scientific Rep 2017). Furthermore, we have incorporated the CRISPR-Cas system, which has made significant advancements in recent years. We have reported protocols for creating deletions across megabase-sized genomic regions (Miyata, DNA Res 2023) and for precise genome editing (Higashitani, Scientific Rep 2023).
2. Analysis of Heterogeneity and Fluctuation Dynamics of Pluripotency of ES Cells
Recent advances in single-cell analysis techniques have revealed that heterogeneity occurs even in seemingly homogeneous cells, that the pluripotency of ES cells varies from cell to cell, and that the state of pluripotency changes dynamically. Understanding the heterogeneity of pluripotency is important not only to reveal the universal principles of life phenomena, but also to efficiently use ES/iPS cells for regenerative medicine. We use fluorescent proteins to visualize the fluctuating state of pluripotency in mouse ES cells, and we analyze the regulatory mechanism of ES cell pluripotency by using fluorescent proteins as indicators. In addition, through comprehensive forward genetic screening, we are currently analyzing previously unknown fluctuations in the pluripotency of ES cells.
Movie 2. Changes in pluripotent state in mouse ES cells
Green: Nanog-EGFP, Red: H2B-mCherry (Nucleus)
3. Elucidation of the Causes of Human Diseases Using Human iPS Cells and Genome Science
We are applying the knowledge we have gained through the aforementioned mouse ES cell research to human iPS cells in order to establish iPS cells derived from human diseases and to construct disease model experimental systems by inducing differentiation of iPS cells. In doing so, we are also conducting comprehensive genome analyses, such as RNA-seq and whole genome sequencing, by using next-generation sequencers.



